The EBMT dataset¤
Data from the European Society for Blood and Marrow Transplantation (EBMT)
A data frame of 2279 patients transplanted at the EBMT between 1985 and 1998.
(from R mstate package, see: https://www.rdocumentation.org/packages/mstate/versions/0.3.1/topics/EBMT%20data)
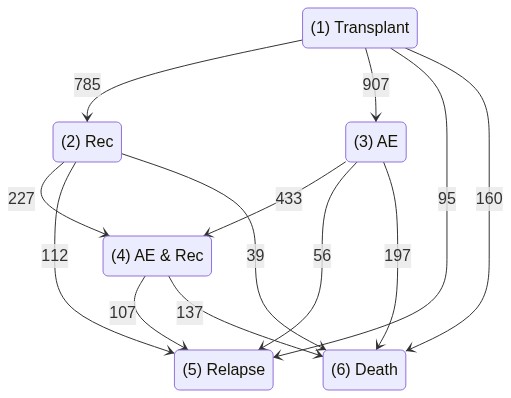
Multi-state model based on Survival-Trees¤
Single patient statistics¤
from pymsm.statistics import prob_visited_states, stats_total_time_at_states
all_states = competing_risk_dataset["target_state"].unique()
# Probability of visiting any of the states
for state in all_states:
if state == 0:
continue
print(
f"Probabilty of {state_labels[state]} = {prob_visited_states(mc_paths, states=[state])}"
)
# Probability of terminal states - Death and Relapse
print(
f"Probabilty of any terminal state = {prob_visited_states(mc_paths, states=multi_state_model.terminal_states)}"
)