PyMSM contains a few example datasets which can easily be loaded:
AID-SI¤
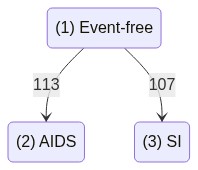
A simple 2 state competing risks dataset
From Data Analysis with Competing Risks and Intermediate States, by Ronald B. Geskus:
The data contains information on two event types, “AIDS” and “SI”, which compete to be the first to occur.
Time is given in years since HIV infection. There are two different representations of the information on the event type that occurred first.
In the cause column, the event type is described in words, whereas in the status column a numeric respresentation is used.
The ccr5 column contains the information on the presence of the deletion CCR5-∆32. Individuals that don’t have the deletion have the value WW (W stands for “wild type”). Individuals that have the deletion on one of the chromosomes have the value WM (M stands for “mutation”)
See also: https://www.rdocumentation.org/packages/mstate/versions/0.3.1/topics/aidssi
Rotterdam¤
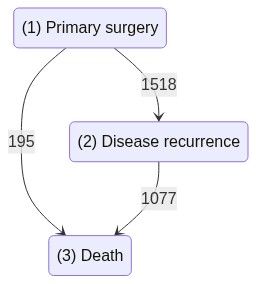
A 3-state Illness-death dataset
The rotterdam data set includes 2982 primary breast cancers patients whose data records were included in the Rotterdam tumor bank. Patients were followed for a time ranging between 1 to 231 months (median 107 months), and outcomes were defined as disease recurrence or death from any cause.
This data includes 2982 patients, with 15 covariates, and was extracted from R survival package.
For more information see page 113 in https://cran.r-project.org/web/packages/survival/survival.pdf.
EBMT¤
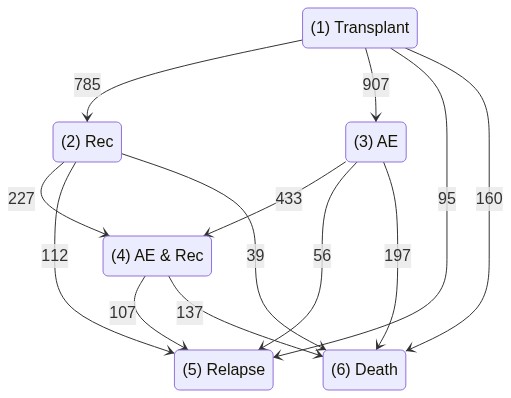
A Multi-state dataset
Data from the European Society for Blood and Marrow Transplantation (EBMT)
A data frame of 2279 patients transplanted at the EBMT between 1985 and 1998.
(from R mstate package, for more information see: https://www.rdocumentation.org/packages/mstate/versions/0.3.1/topics/EBMT%20data)
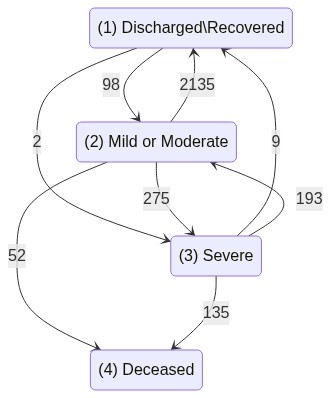
Covid hospitalizations in Israel¤
Complex multis-state, recurring events and time-varying covariates dataset
Israel COVID-19 hospitalization public data, as described in Roimi et. al. 2021.