Multi-State Model - rotterdam example¤
In this notebook¤
This notebook provides a first example of using the pymsm package, using the rotterdam dataset.
Rotterdam dataset¤
The rotterdam data set includes 2982 primary breast cancers patients whose data records were included in the Rotterdam tumor bank. Patients were followed for a time ranging between 1 to 231 months (median 107 months), and outcomes were defined as disease recurrence or death from any cause.
This data includes 2982 patients, with 15 covariates, and was extracted from R survival package.
For more information see page 113 in https://cran.r-project.org/web/packages/survival/survival.pdf.
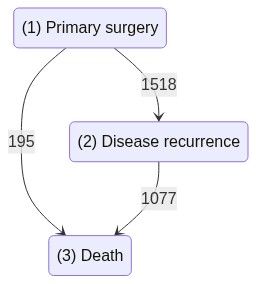
Let’s load the dataset, which holds the transitions for each patient between the three states as decribed in the graph below
The dataset is a list of elements from class PathObject. Each PathObject in the list corresponds to a single sample’s (i.e “patient’s”) observed path.
Let’s look at one such object in detail:
We see the following attributes:
-
sample_id : (optional) a unique identifier of the patient.
-
states : These are the observed states the sample visited, encoded as positive integers. Here we can see the patient moved from state 1 to 2, ending with the only terminal state (state 3).
-
time_at_each_state : These are the observed times spent at each state.
-
covariates : These are the patient’s covariates
- “year”
- "age"
- "meno"
- "grade"
- "nodes"
- "pge"
- "er"
- "hormone"
- "chemo"
Note: if the last state is a terminal state, then the vector of times should be shorter than the vector of states by 1. Conversely, if the last state is not a terminal state, then the length of the vector of times should be the same as that of the states. In such a case, the sample is inferred to be right censored.
Updating Covariates Over Time¤
In order to update the patient covariates over time, we need to define a state-transition function. In this simple case, the covariates do not change and the function is trivial
You can define any function, as long as it recieves the following parameter types (in this order):
1. pandas Series (sample covariates when entering the origin state)
2. int (origin state number)
3. int (target state number)
4. float (time spent at origin state)
5. float (absolute time of entry to target state)
If some of the parameters are not used in the function, use a default value of None, as in the example above.
Defining terminal states¤
Fitting the model¤
Import and init the model
Fit the model
Making predictions¤
Predictions are done via monte carlo simulation. Initial patient covariates, along with the patient’s current state are supplied. The next states are sequentially sampled via the model parameters. The process concludes when the patient arrives at a terminal state or the number of transitions exceeds the specified maximum.
import numpy as np
all_mcs = multi_state_model.run_monte_carlo_simulation(
# the current covariates of the patient.
# especially important to use updated covariates in case of
# time varying covariates along with a prediction from a point in time
# during hospitalization
sample_covariates=dataset[0].covariates.values,
# in this setting samples start at state 1, but
# in general this can be any non-terminal state which
# then serves as the simulation starting point
origin_state=1,
# in this setting we start predictions from time 0, but
# predictions can be made from any point in time during the
# patient's trajectory
current_time=0,
# If there is an observed upper limit on the number of transitions, we recommend
# setting this value to that limit in order to prevent generation of outlier paths
max_transitions=2,
# the number of paths to simulate:
n_random_samples=100,
)
The Simulation Results Format¤
Each run is described by a list of states and times spent at each state (same format as the dataset the model is fit to).
Below are two samples: